Review: LAT Data Analysis Compare: pyLikelihood - Unbinned and Binned Analysis Perform:
Example: gt_apps python Script Used in Binaries RSP Analysis; (For more examples, see Richard Dubois' confluence page "Scripts used in Binaries RSP Analysis". See pyLikelihood: Also see: |
pyLikelihood: BinnedAnalysis
Prerequisites
Procedure (SCons ScienceTools-09-20-00)- Log in to SLAC Public and change to your work directory (e.g., mypyLike/binned_analysis).
Note: If you are running on a Windows machine, use X-Win32 as your X server.
- Set up your environment; see Setup: SLAC Central Linux.
Note: Be sure to use a bash shell.
- Using your editor, create a simple python script file to perform an BinnedAnalysis and name it (e.g., my_binned_analysis.py; see example).
- From the command line, enter:
python
- From the python prompt, enter:
from my_binned_analysis import *
If you see the error message "Caught ImportError: libtk8.4.so: cannot open shared object file: No such file or directory", ignore it.
Your pyLikelihood BinnedAnalysis results will be displayed. (Note that the fit is: 107731.417122.)
- From the python prompt, print the plots:
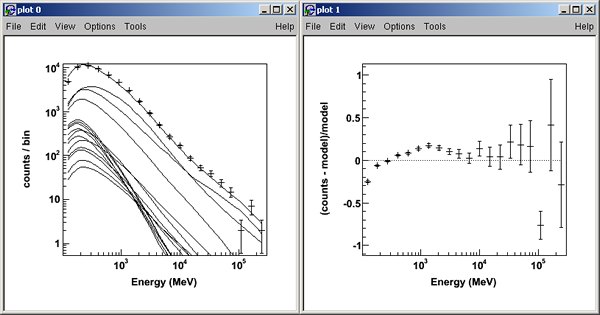
>>> like.plot()
Note: By default, the plot() method uses pyROOT to create counts spectra plots for comparing the model fits to the data. The two plot windows created are shown below; one for the counts spectra, and one for the fractional residuals of the fit.
- Challenge: You may want to review Model Fitting with gtlikelihood and then perform the analysis again and try to improve the fit.
| Owned by: | Jim Chiang |
| Last updated by: Chuck Patterson 04/01/2011 |