Analyzing Full GLAST ROOT Trees
This section provides the following detailed examples of how to perform a RootTreeAnalysis:
Prerequisites
To analyze the full ROOT trees, you will need:
- ROOT installed for standalone use as an analysis tool. (See Install ROOT.)
- A copy of RootTreeAnalysis available in the RootAnalysis package and also distributed with EngineeringModel checkout package.
Tip: Place these files in the same directory (e.g., yourWorkDirectory) from which you launch ROOT.
Note: A current copy as presented in this tutorial is available for download at:
- GLAST ROOT Offline Libraries: commonRootData, mcRootData, digiRootData, reconRootData (required for reading the full ROOT trees); the libraries for linux and windows can be obtained using the GLAST Installer.
(See Install ROOT.)
| Note: When downloading libraries, be sure to use the version that were used to create the data you are analyzing. Due to the reorganization of the TkrRecon data, not all ROOT files are readable by all versions of the libraries. |
Full documentation of the GLAST ROOT data classes is available for:
Introduction
In addition to the Summary Ntuple, four full ROOT trees are potentially available as output from Gleam: monte carlo, digitization, reconstruction, relational table. Analyzing the data in these files is more complex due to the more complicated GLAST-defined data structures in use to store the data. Often, it is possible to merely use the summary ntuple for simple analysis. However, if one is interested in details of an event, it may be necessary to look at the full ROOT trees.
GLAST Offline provides RootTreeAnalysis as an example ROOT macro that can be used to read Digitization, Reconstruction, and/or Monte Carlo GLAST ROOT files. Output is a new ROOT file (named Histograms.root by default) which contains a series of user-defined histograms. Users can provide individual ROOT files as input, or TChains of files for multi-file processing.
Defining histograms, ntuples and accessing those objects to fill them.
Event processing is done by calling RootTreeAnalysis::Go(Int_t numEvents); This causes the next numEvents in the files to be processed - filling the defined histograms.
Note: This does not cause the histograms to be saved automatically!! The user must call RootTreeAnalysis::WriteHist() to save the histograms.
The actual analysis is performed in a series of routines called by Go( ). The methods that handle the digitization data are called: DigiTkr( ), DigiCal( ), and DigiAcd( ). Similarly, the reconstruction methods are: ReconTkr( ), ReconCal( ). When adding your own analysis - you may add it directly to the Go( ) method - or you may prefer to create your own new method that specifically handles the type of data you desire to access. In addition, you may use the predefined histograms provided or modify/add/remove histograms. Users define their histograms in the HistDefine methods, located in the RootTreeAnalysis.cxx file.
The following files describe how to access data from four specific categories:
TKR digitization
TKR reconstruction
CAL digitization
CAL reconstruction
Example 1: RootTreeAnalysis Reading in Single
mc, digi, and/or recon ROOT files:
Notes: Be sure to:
- Change to your work directory (i.e., the directory containing all of the files downloaded for this analysis) before launching ROOT.
|
- Start up ROOT and, at the root [ ] prompts, load the GLAST ROOT
libraries - in the order shown - using the following commands:
Linux |
Windows |
| gSystem->Load("libcommonRootData.so") |
gSystem->Load("commonRootData.dll") |
| gSystem->Load("libmcRootData.so") |
gSystem->Load("mcRootData.dll") |
| gSystem->Load("libdigiRootData.so") |
gSystem->Load("digiRootData.dll") |
| gSystem->Load("libreconRootData.so") |
gSystem->Load("reconRootData.dll") |
Notes: The Linux libcommonRootData.so (or Windows commonRootData.dll) file should be loaded first, since it is required by the other GLAST ROOT libraries; otherwise, you will receive a warning message about undefined classes.
- Load the RootTreeAnalysis macro:
.L RootTreeAnalysis.cxx
- Create a new instance of RootTreeAnalysis that includes the files you wish to analyze:
RootTreeAnalysis *r = new RootTreeAnalysis("digi.root", "recon.root", "mc.root")
- Loop over 6 events:
r->Go(6)
The response will be similar to: Num Event in File is: 40000.
Note: If you get too few events, increase the number of "loop over" events; if you get a message re: "Interpreter error recovered", decrease the number.
- Save the Histograms to a file:
r->WriteHist()
Note: By default this file is named Histograms.root.
- Start up a TBrowser:
TBrowser tb
- View the TKRSTRIPSLYR5 histogram using the TBrowser window.
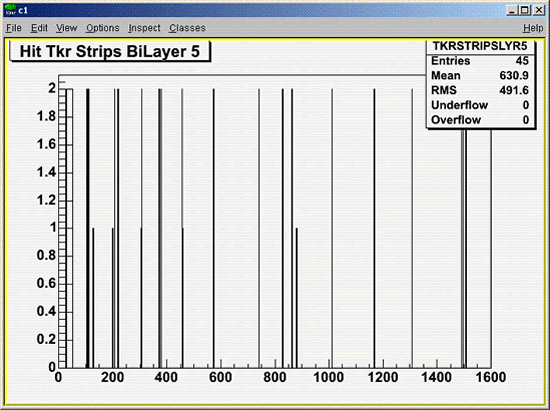
- To quit Root, enter:
.q
Example 2: RootTreeAnalysis Reading in a
TChain of mc, digi, and/or recon ROOT files
Notes: Be sure that you:
- Change to your work directory (i.e., the directory containing all of the files downloaded for this analysis) before launching ROOT.
- Start up ROOT and, at the root [ ] prompts, load the GLAST ROOT libraries - in the order shown - using the following commands:
Linux |
Windows |
| gSystem->Load("libcommonRootData.so") |
gSystem->Load("commonRootData.dll") |
| gSystem->Load("libmcRootData.so") |
gSystem->Load("mcRootData.dll") |
| gSystem->Load("libdigiRootData.so") |
gSystem->Load("digiRootData.dll") |
| gSystem->Load("libreconRootData.so") |
gSystem->Load("reconRootData.dll") |
Note: The libcommonRootData.so or .dll file should be loaded first, since it is required by the other GLAST ROOT libraries; otherwise, you will receive a warning message about undefined classes.
- Create a TChain of ROOT files for each type of ROOT file;
one TChain each for: mc, digi, and recon:
TChain *mcChain = new TChain("Mc")
TChain *digiChain = new TChain("Digi")
TChain *recChain = new TChain("Recon")
Note: When creating the TChain, you pass in - as a parameter - the name of the TTree; in this case: Mc, Digi, or Recon.
- Add the desired files to each TChain:
mcChain->Add("mc*.root")
digiChain->Add("digi*.root")
recChain->Add("recon*.root")
Note: When calling TChain::Add, you can use wildcards to add more than one ROOT file to the chain.
- Load the RootTreeAnalysis macro:
.L RootTreeAnalysis.cxx
- Create a new instance of RootTreeAnalysis that includes the chains you wish to analyze:
RootTreeAnalysis *r = new RootTreeAnalysis(digiChain, recChain, mcChain)
- Loop over six events:
r->Go(6)
- Save the Histograms to a file:
r->WriteHist()
Note: By default this file is named Histograms.root.
- Start up a TBrowser:
TBrowser tb
- View some histograms using the TBrowser window.
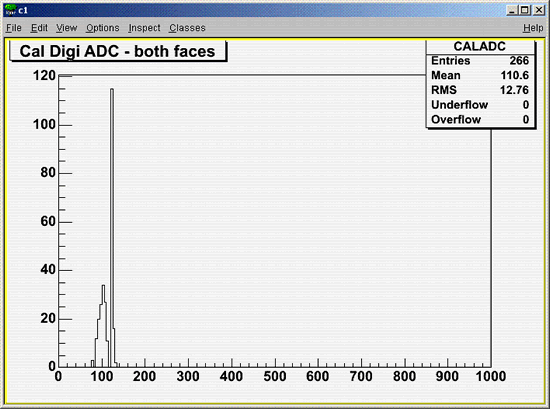
- To quit Root, enter:
.q
References:
- RootTalk Forum and RootTalk Digest, available from the ROOT home page.
Notes:
Also see:
Owned by: Heather Kelly
| Last updated by: Chuck Patterson
07/14/2009 |
|
|